Atomistic graph analysis of relevant proteins of SARS-CoV-2
The current global pandemic of COVID-19 has brought the causing pathogen family of coronaviruses back to the centre of attention. These viruses have already been linked to pandemic diseases, like the severe acute respiratory syndrome (SARS) in 2003 [13] and the Middle East respiratory syndrome (MERS) in 2012 [14]. The current disease, COVID-19, is responsible for over 26 million infections globally and over 850 thousand deaths (as of 01.09.2020) [source: WHO https://covid19.who.int]. The responsible virus has been termed SARS-CoV-2 as it has an 82% genomic similarity with the SARS coronavirus [15,16]. Proteins of these viruses have aready been the focus of intense research to aid in drug discovery but the current situation has provided renewed urgency. Here, we use the theoretical methodologies underpinning ProteinLens to investigate the allosteric properties of the SARS-CoV-2 main protease Mpro (also called 3CL protease). We also give direct access to the results via interactive ProteinLens output pages. For detailed information, click on the box below.
Inhibiting the main protease (Mpro) of SARS-CoV-2 is of great interest in addressing the COVID-19 disease. The protein is essential for virus replication and drugs disrupting this process may provide an efficient treatment for COVID-19 patients. Although most efforts are centred on inhibiting the binding site, we report here on the allosteric communication pathways in the Mpro dimer as obtained by Bond-to-bond propensity and Markov transient analysis. When sourced from the active site (green), we obtain the allosteric properties of each bond and atom, from cold (blue scale) to hot (red scale) and further score residues to identify those that mediate interactions through the whole protein. You can interactively explore the figures below to see how the main protease behaves under allosteric regulation at the atomistic level through Bond-to-bond propensities and can find more details on this ProteinLens results page: NJMSBB.
We further score the sites of interest for significance by using the average residue quantile score and contrast them against randomly sampled sites (read more here). This allowed us to identify four putative allosteric hotspots on the protease as attractive alternative drug targets to the main binding site of the enzyme. Two of these sites identified with Bond-to-bond propensities are shown in the Figure below.
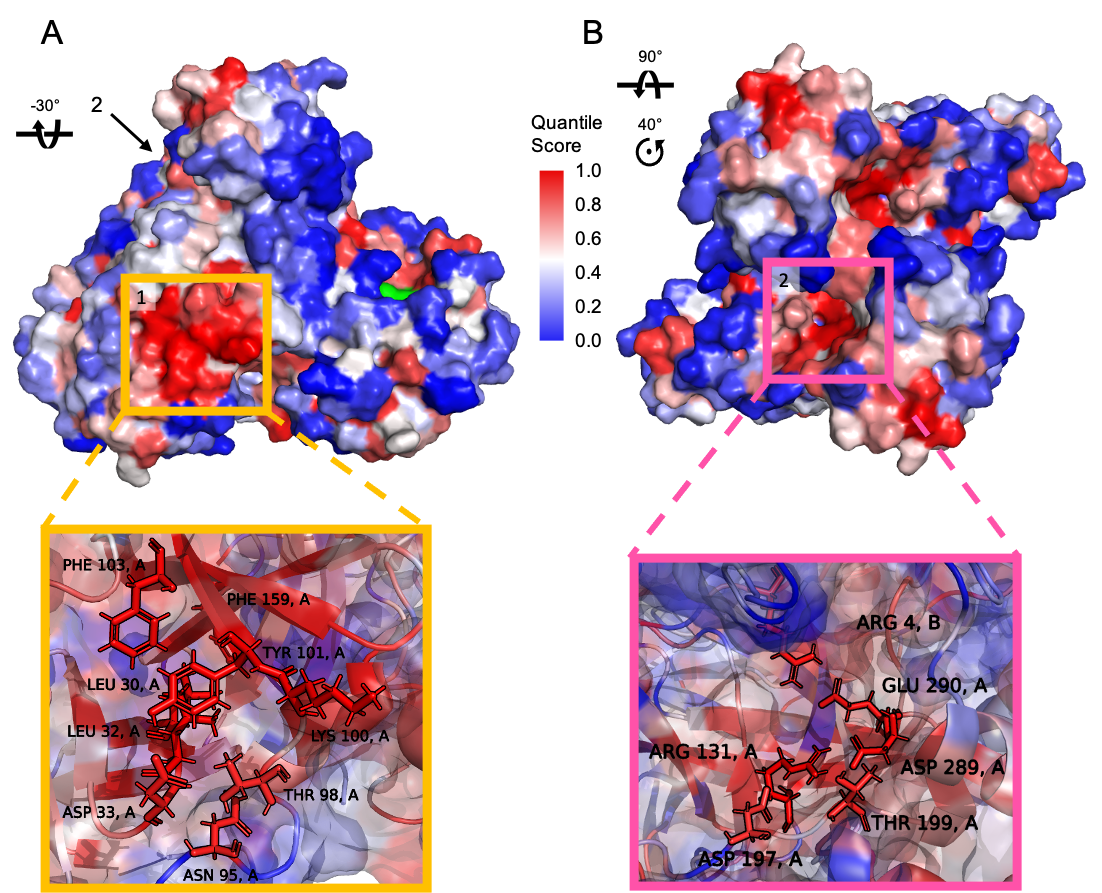
To explore our findings on the main protease interactively, you can follow this session ID: NJMSBB, which links to the Bond-to-bond propensity as well as Markov Transient analyses and further scoring results. For further details and a full list of residues in all four hotspots (only two of which are shown here), please also see the following preprint: Strömich, L., Wu, N., Barahona, M., & Yaliraki, S. N. (2020). Allosteric Hotspots in the Main Protease of SARS-CoV-2. BioRxiv, DOI: 2020.11.06.369439 (currently under review).