Welcome to ProteinLens
ProteinLens constructs a fully atomistic, energy-weighted graph representation of a biomolecular structure and explores the long-range communication or connectivity between any specific sites in the system. For example, it can be used to:
- identify potential allosteric sites
- explore allosteric pathways
- identify residues that play a key role in signalling and/or cooperativity
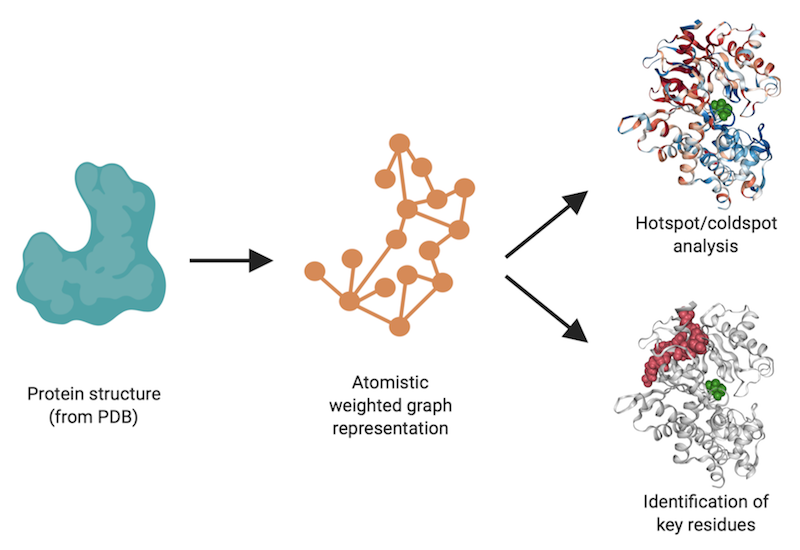
The Background page provides insights into the theory behind ProteinLens, along with some recent examples of our work using the here presented approaches. This web-based application allows the construction and download of the graph corresponding to the structure of your choice, the interactive visualisation of residues relevant for communication within or across proteins and the download of complete results files. Visit the Tutorial page for a walk-through of the web interface, and the FAQ page if you get stuck!
Start here
with a new session.
- Share - copy and redistribute the material in any medium or format
- Adapt - remix, transform and build upon the material
- Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
- NonCommercial — You may not use the material for commercial purposes.
- No additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits.